
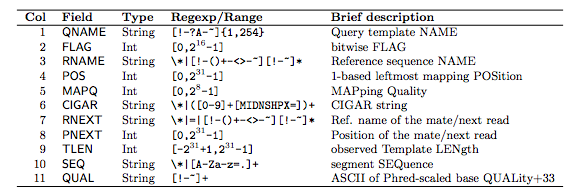
Template: The DNA fragment that was measured Each alignment line/record has 11 mandatory fields describing essential alignment information. The alignment records constitute the body of the file. Header hold generic information on SAM file along with version information, if the file is sorted, information on reference sequence, etc. Header lines start with while alignment lines do not. It is TAB-delimited text format with header and a body. SAM format files are generated following mapping of the reads to reference sequence. Most often it is generated as a human readable version of its sister BAM format, which stores the same data in a compressed, indexed, binary form.
Bam file format series#
The SAM Format is a text format for storing sequence data in a series of tab delimited ASCII columns. It is critical to figure out the PHRED score type used in. The figure below illustrates the PHRED usage across different sequencing notations. on the base character ( character that represents zero PHRED score ), PHRED scale is often referred as FHRED+33 (ASCII character !) or FHRED+64 (ASCII Character ?). Why 33 to 126? Because 33 to 126 codes for single characters, so the score can be represented by a single character. Rather than giving numeric values of PHRED score they are provided in ASCII character codes from 33 to 126. Probability that the base is called wrongįastq-sanger holds PHRED score from 0-93 whereas fastq-Illumina provides PHRED scores from 0-62.
Where p is the probability that the corresponding base call is incorrect. It indicates how confident we can be that the base was sequenced and identified correctly. Line 4: AAAFFJJJJJJJJJJJJJJJJJFJJFJJJJJFJJJJJJJJJJJJJJJJ#FJ#JJJJF#F#FJJ#F#JJJFJJJJJĪ quality score ( PHRED scale) for each base pair. Line 2: ATAATAGGATCCCTTTTCCTGGAGCTGCCTTTAGGTAATGTAGTATCTNATNGACTGNCNCCANANGGCTAAAGT

Lines with ‘ ’ are not a common feature of fasta files. Any other line that starts with ‘ ’ will be ignored. First line referred as comment line starts with ‘>’ and gives basic information about sequence. This is a very basic format with two minimum lines. TGGCTGTGATGGCTTTTAGCGGAAGCGCGCTGTTCGCGTACCTGCTGTTTGTTGAAAATTTAAGAGCAAAGTGTCCGGCTCGATCCCTGCGAATTGAATTCTGAACGCTAGAGTAATCAGTGTCTTTCAAGTTCTGGTAATGTTTAGCATAACCACTGGAGGGAAGCAATTCAGCACAGTAATGCTAATCGTGGTGGAGGCGAATCCGGATGGCACCTTGTTTGTTGATAAATAGTGCGGTATCTAGTGTTGCAACTCTATTTTTįasta format is a simple way of representing nucleotide or amino acid sequences of nucleic acids and proteins.
Bam file format manual#
Please refer user manual or other information resources on web for more details.įile extensions : file.fa, file.fasta, file.fsaĮxample : >XR_002086427.1 Candida albicans SC5314 uncharacterized ncRNA (SCR1), ncRNA The information provided here is basic and designed to help users to distinguish the difference between different formats. This section explains some of the commonly used file formats in bioinformatics.


 0 kommentar(er)
0 kommentar(er)
